ITS2 CBC species concept - A Pandora´s box of protist systematics
Species represent basic units of taxonomic classification, and play a fundamental role in assessing diversity of Life. In protists, a method of species delineation commonly used in recent years is based on the identification of Compensatory Base Changes1 (CBCs) in the secondary structure of the ITS2 molecule (the spacer between the 5.8S and 28S rRNA genes).
The ITS2 CBC – based species concept involves two main approaches: 1) sensu Coleman2 [1, 2] and 2) sensu Müller et al.3 [3]. These two approaches, although differing in detail, fully rest on the homology of the investigated ITS2 positions. Therefore their applicability and reproducibility requires a robust prediction of the ITS2 secondary structure of the organisms under study.
In our laboratory we have studied the evolution of the ITS2 molecule in several lineages of green algae (Chaetophorales, Chaetopeltidales, Oedogoniales, Sphaeropleales, and Ulvales,) with the primary focus on CBCs in the conserved regions of their ITS2 molecule. As a result we found that CBCs are not diagnostic at the species level, and that even genera, families and orders could lack CBCs in such regions (see fig. 2 in Caisová et al. [4] and fig. 1 in Caisová et al. [5]).
Since our findings are largely incongruent with the basic assumption of the ITS2 CBC – based species concept (i.e. the correlation between CBCs in ITS2 and recent speciation events), we have evaluated each step in the process starting from the secondary structure prediction and ending with the identification of CBCs among the studied organisms. Interestingly, we found that the currently used criteria/landmarks4 for folding (employed also in automatic tools for secondary structure prediction) are incompatible with the phylogenetically derived secondary structure. They mostly fail to identify the basal positions of the ITS2 Helices, i.e. the helix boundaries, which leads to false homology assessments of base pairing in the predicted secondary structures. This often invalidates the identification of CBCs and thus of species based on spurious CBCs.
Recently, we presented:
1) the first ITS2 consensus secondary structure model that can be applied to two classes of green algae (Chlorophyceae and Ulvophyceae – Ulvales), [4, 5]
2) new landmarks (the spacer regions separating the ITS2 Helices, for robust prediction of the ITS2 secondary structures in general, for details see Fig. 1.
Since the ITS2 molecule has been widely used in molecular phylogeny of protists, in barcoding projects [e.g. 6, 7] and for identification of novel species of diverse groups of organisms (mostly algae), we proposed a general strategy of how to obtain a robust ITS2 secondary structure for any group of organisms [5].
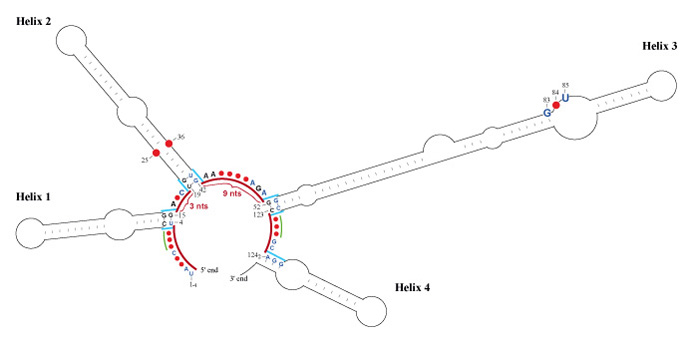
Fig. 1. General model of the ITS2 concensus secondary structure in two classes of green algae – Chlorophyceae and Ulvophyceae (Ulvales). The common core ITS2 secondary structure consists of four usually unbranched helices and spacers between helices. The unique spacers (red lines and red parentheses accompanied by number of nucleotides) specify the boundaries of helices, except in the highly variable 3' end of Helix 4. The basal parts of helices (blue lines) are characterized by their first two base pairs (Helices 1, 2, 3) or by three nucleotides on the 5' side (Helix 4). The variability map shows the degree of conservation of spacers and the basal parts of helices (for variability categories, see [5]). The universal numbers define 1) the starting points of the helices and 2) the common motifs in Helix 2 (pyrimidine-pyrimidine mismatch: 25/36) and Helix 3 (GYU: 83 – 85). The proposal is based on 252 sequences of green algae [105 Ulvales (Ulvophyceae); 31 Chaetophorales, 3 Chaetopeltidales, 18 Oedogoniales, and 95 Sphaeropleales (Chlorophyceae)].
Glossary:
1Compensatory Base Change: CBC, a double-sided base change of a nucleotide pair in a helix, retaining the secondary structure [8]
2The ITS2 CBC species concept sensu Coleman is based on a correlation found between the presence/absence of CBCs in conserved regions of the ITS2 secondary structure (Helix 2 and 3) and the inability/ability of strains to sexually cross. A CBC involving the conserved regions of Helices 2 and 3 indicated inability of strains to sexually cross [1].
3The ITS2 CBC species concept sensu Müller et al. is based on a correlation found between CBCs in the whole ITS2 molecule (four helices including their variable parts) and organisms classified as different species (mostly plants and fungi). According to Müller and colleagues (2007) [3].
4criteria/landmarks: the currently used criteria for folding are 1) the presence of (mainly) four helices with Helix 3 as the longest one, 2) a pyrimidine-pyrimidine mismatch in Helix 2, and 3) a YGGY motif (or its derivatives) on the 5' side of Helix 3 [9, 10]
References:
- Coleman AW (2000) The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist 151: 1–9
- Coleman AW (2009) Is there a molecular key to the level of "biological species" in eukaryotes? A DNA guide. Mol Phylogenet Evol 50: 197–203
- Müller T, Philippi N, Dandekar T, Schultz J, Wolf M (2007) Distinguishing species. RNA 13: 1469–1472
- Caisová L, Marin B, Melkonian M (2011) A close-up view on ITS2 evolution and speciation - a case study in the Ulvophyceae (Chlorophyta, Viridiplantae). BMC Evol Biol 11: 262
- Caisová, L., Marin, B., Melkonian, M. (2013): A Consensus Secondary Structure of ITS2 in the Chlorophyta Identified by Phylogenetic Reconstruction. Protist 164: 482-496 doi:10.1016/j.protis.2013.04.005
- Stech M, Veldman S, Larraín J, Muñoz J, Quandt D, Hassel K, Kruijer H (2013) Molecular Species Delimitation in the Racomitrium canescens Complex (Grimmiaceae) and Implications for DNA Barcoding of Species Complexes in Mosses. PLoS ONE 8(1): e53134
- Yao H, Song J, Liu C, Luo K, Han J, Li Y, Pang X, Xu H, Zhu Y, Xiao P, Chen S (2010) Use of ITS2 Region as the Universal DNA Barcode for Plants and Animals. PLoS ONE 5(10): e13102
- Gutell RR, Larsen L, Woese CR (1994) Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev 58: 10–26
- Mai JC, Coleman AW (1997) The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. J Mol Evol 44: 258–271
- Schultz J, Maisel S, Gerlach D, Muller T, Wolf M (2005) A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 11:361–364
Contact: Lenka Caisová
|