|
|
Paulinella chromatophora – A model organism for acquisition of photosynthesis by eukaryotes
One key step in the evolution of life was the origin of photosynthesis in nucleated cells. This process was initiated when a cyanobacterium was engulfed by a colourless host-cell more than a billion years ago and transformed into a plastid during a process known as primary endosymbiosis. It is commonly accepted that a single primary endosymbiosis gave rise to all plastids.
Recently, we presented evidence that photosynthetic inclusions, termed chromatophores, present in the filose thecamoeba Paulinella chromatophora (Fig. 1) originated from an independent, more recent primary endosymbiotic event (Fig. 2, and [1, 2, 3]). To clarify metabolic capabilities of the chromatophore, and its state of integration into the host, we analysed the complete genome sequence of the chromatophore in collaboration with Dr. Gernot Glöckner from the Fritz-Lipmann-Institute (Jena).
|
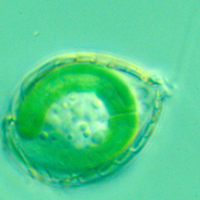
Fig1
|
Our study revealed a fundamental reduction of the chromatophore genome [4]. The single circular chromosome of 1.02 Mb encodes 867 protein-coding genes and is therewith the smallest cyanobacterial genome reported to date. Compared to Synechococcus WH5701, a free-living relative of the chromatophore, only 26% of the genes were retained. 11 putative pseudogenes were identified, indicating that reductive genome evolution is ongoing. While the chromatophore genome contains a complete set of photosynthesis genes, it lacks not only genes thought to be dispensable for an intracellular lifestyle, but also genes of essential pathways for amino acid and co-factor synthesis.
Our study characterized the chromatophore as a photosynthetic entity that is absolutely dependent on its host for growth and survival. Thus, the chromatophores of P. chromatophora are the only known cyanobacterial descendants, besides plastids, with a significantly reduced genome that confer photosynthesis to their eukaryotic host. Their comparison with plastids and bacterial endosymbionts of invertebrates sheds light on early steps of the integration of a photosynthetic prokaryote into a eukaryotic cell.
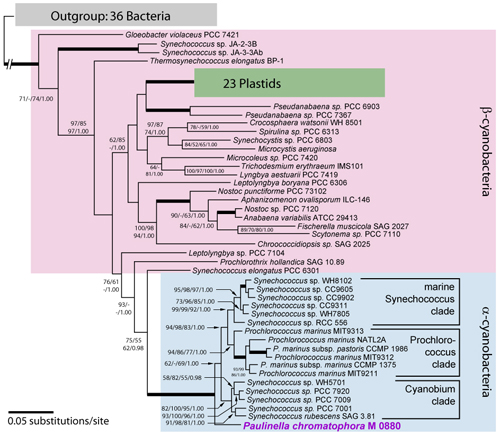
Fig. 2. Phylogenetic evidence for an independent cyanobacterial origin of the chromatophore of Paulinella chromatophora, significantly separated from the plastid lineage. The maximum-likelihood tree (GTR+I+G) is based on rDNA operon sequences (genes encoding SSU rRNA, tRNA-Ile, tRNA-Ala, and LSU rRNA); numbers at branches are bootstrap values [maximum likelihood, neighbor joining and maximum parsimony (>50%)] and Bayesian posterior probabilities >0.95 (branches in bold have maximum support).
References:
- Marin B, Nowack ECM, Melkonian M: A plastid in the making: Evidence for a second primary endosymbiosis. Protist 2005, 156(4):425-432.
- Marin B, Nowack ECM, Glöckner G, and Melkonian M: The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus-like gamma-proteobacterium. BMC Evol Biol 2007, 7:85.
- http://www.faz.net
- Nowack ECM, Melkonian M, and Glöckner G: Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 2008, 14(6):410-418.
|
|